News from the Omar Lab
New Preprint: THR-6E for Risk Stratification in ER+/HER2− Breast Cancer
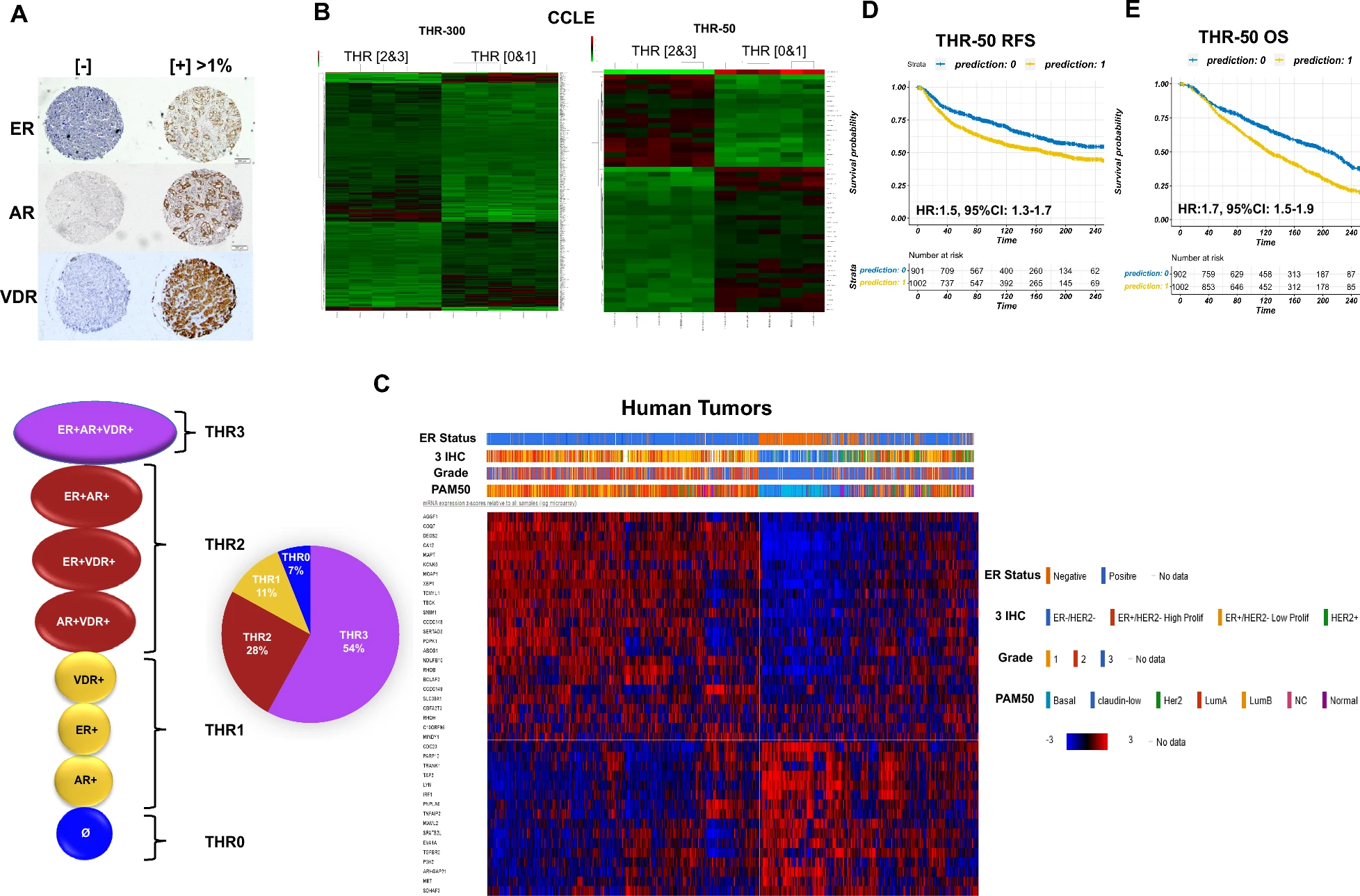
We are excited to share our new preprint, “THR-6E: A Six-Gene Cell-of-Origin Signature Stratifies Risk and Predicts Systemic Therapy Response in ER+/HER2− Breast Cancer,” now available on medRxiv.
Building on our earlier work on the triple hormone receptor (THR) signature, THR-6E is a compact six-gene cell-of-origin signature (KIF4A, KIF2C, CDC20, FAM64A, TPX2, LMNB2) that robustly stratifies relapse-free survival and predicts response to both endocrine therapy and chemotherapy in ER+/HER2− breast cancer. The signature was validated across >7,000 breast cancer cases from multiple independent cohorts, including the I-SPY2 adaptive clinical trial.
Notably, THR-6E maintains its prognostic and predictive value in lymph node-positive patients — a subgroup where existing genomic assays such as Oncotype DX, MammaPrint, and Prosigna have limited clinical utility and are not yet strongly recommended by current guidelines. THR-6E predicted chemotherapy response in node-positive ER+/HER2− disease with ~70% sensitivity and specificity across taxane-, anthracycline-, and FEC-based regimens, addressing a key unmet need in this patient population.
Omar Lab Presents at the NRG Oncology 2026 Winter Meeting
Dr. Mohamed Omar presented at the NRG Oncology 2026 Winter Meeting in San Francisco, delivering two talks on THR-based signatures for forecasting clinical outcome and treatment response in breast cancer.
The first presentation was to the Pathology Committee, and the second to the Translational Science Committee, both focusing on our work developing cell-of-origin signatures for risk stratification and therapy response prediction in breast cancer patients.
These presentations highlight ongoing collaborations within NRG Oncology to advance precision approaches for breast cancer management.
New Subaward to Advance Multi-Omics Integration
We are pleased to announce that the Omar Lab has received a subaward from the National Institutes of Health and City University of New York (CUNY) to support the development of cross-platform analytical frameworks for multi-omics integration and digital pathology.
The two-year project, titled “Multi-Omics Integration of TCGA Histopathology Images through a Cross-Platform Pipeline Bridging Python and R Bioconductor,” aims to create a robust computational infrastructure for integrating histopathology-derived image features with genomic, transcriptomic, and clinical data from large cancer cohorts such as TCGA.
By bridging Python-based deep learning pipelines with R/Bioconductor statistical environments, this project will streamline multimodal data fusion and reproducibility, enabling researchers to explore tumor biology across molecular and morphological scales.
This collaboration further advances our mission to develop open and interoperable frameworks for multimodal cancer research, combining AI-powered pathology with rigorous multi-omics integration.

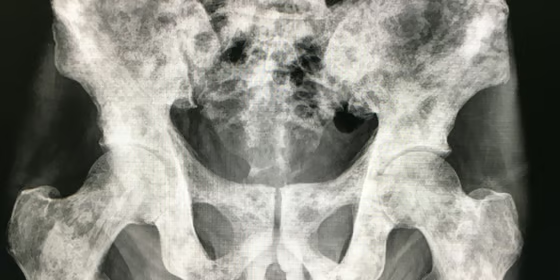
New DoD Award to Decode Bone Metastasis in Prostate Cancer
We are excited to share that the Omar Lab has received a U.S. Department of Defense Prostate Cancer Research Program (PCRP) Idea Development Award to support our ongoing efforts to understand and predict bone metastasis in prostate cancer.
This three-year project, titled “Decoding Bone Metastasis in Prostate Cancer: A Multimodal Approach Integrating Genomics and Imaging Data,” aims to uncover the molecular and spatial mechanisms that allow prostate cancer cells to colonize bone and drive disease progression. Our team will develop integrative computational frameworks that combine genomic, transcriptomic, and imaging data to identify early predictors of metastasis directly from diagnostic tumor samples.
Bone metastasis remains the leading cause of death among men with prostate cancer, often resulting in fractures, severe pain, and loss of mobility. By identifying the molecular and architectural features that predispose tumors to spread, our work seeks to enable personalized treatment strategies — offering aggressive therapy for high-risk patients while sparing low-risk individuals from unnecessary intervention.
This award will accelerate our ongoing efforts to integrate artificial intelligence, digital pathology, and multi-omics data to uncover the mechanisms that govern tumor–bone microenvironment interactions in prostate cancer.
The Omar Lab is Now Open at Cedars-Sinai!
We are excited to announce that the Omar Lab is now open at Cedars-Sinai Medical Center!!. We are part of the Department of Computational Biomedicine and the Samuel Oschin Comprehensive Cancer Institute We are a hybrid lab with specific interest in developing AI-powered risk assessment and prediction tools for cancer patients informed by the composition of the tumor microenvironment. Check our join page for open positions and opportunities to join our research in sunny LA!

